-Search query
-Search result
Showing all 29 items for (author: battisti & aj)

EMDB-5448: 
Cryo-tomography and subtomogram averaging of the Newcastle disease virus matrix protein array
Method: subtomogram averaging / : Battisti AJ, Meng G, Winkler DC, McGinnes LW, Plevka P, Steven AC, Morrison TG, Rossmann MG
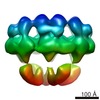
EMDB-2056: 
Electron cryo-microscopy of the Hantaan virus glycoprotein spike
Method: single particle / : Battisti AJ, Chu YK, Chipman PR, Kaufmann B, Jonsson CB, Rossmann MG

EMDB-5106: 
Fab from MAb B interacting with feline panleukopenia virus
Method: single particle / : Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG

EMDB-5107: 
Feline panleukopenia virus in complex with FAb from neutralizing antibody MAb 6
Method: single particle / : Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG

EMDB-5108: 
Feline panleukopenia virus in complex with FAb from neutralizing antibody MAb 8
Method: single particle / : Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG

EMDB-5109: 
Feline panleukopenia virus in complex with FAb from neutralizing antibody MAb 15
Method: single particle / : Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG

EMDB-5110: 
Feline panleukopenia virus in complex with FAb from neutralizing antibody MAb 16
Method: single particle / : Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG

EMDB-5111: 
Feline panleukopenia virus in complex with FAb from neutralizing antibody MAb E
Method: single particle / : Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG

EMDB-5112: 
Feline panleukopenia virus in complex with FAb from neutralizing antibody MAb F
Method: single particle / : Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG

EMDB-1597: 
5-fold averaged density map of Paramecium bursaria Chlorella virus-1 (PBCV-1).
Method: single particle / : Cherrier MV, Kostyuchenko VA, Xiao C, Bowman VD, Battisti AJ, Yan X, Chipman PR, Baker TS, Van Etten JL, Rossmann MG

EMDB-5105: 
Canine parvovirus in complex with FAb from neutralizing antibody MAb 14
Method: single particle / : Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG

EMDB-1580: 
CryoEM and 3D image reconstruction of Chilo Iridescent virus at 13 angstrom resolution
Method: single particle / : Yan X, Yu Z, Zhang P, Battisti AJ, Holdaway HA, Chipman PR, Bajaj C, Bergoin M, Rossmann MG, Baker TS

EMDB-1418: 
Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins
Method: single particle / : Lok SM

EMDB-1280: 
Cryo-electron microscopy study of bacteriophage T4 displaying anthrax toxin proteins.
Method: single particle / : Fokine A, Bowman VD, Battisti AJ, Li Q, Chipman PR, Rao VB, Rossmann MG

EMDB-1266: 
Structural changes of bacteriophage phi29 upon DNA packaging and release.
Method: single particle / : Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG

EMDB-1393: 
Cryo-EM study of the Pseudomonas bacteriophage phiKZ.
Method: single particle / : Fokine A, Battisti A, Bowman V, Efimov A, Kurochkina L, Chipman P, Mesyanzhinov V, Rossmann M

EMDB-1392: 
A three-dimensional cryo-electron microscopy structure of the bacteriophage phiKZ head.
Method: single particle / : Fokine A, Kostyuchenko V, Efimov A, Kurochkina L, Sykilinda N, Robben J, Volckaert G, Hoenger A, Chipman P, Battisti A, Rossmann M, Mesyanzhinov V

EMDB-1415: 
Cryo-EM study of the Pseudomonas bacteriophage phiKZ.
Method: single particle / : Fokine A, Battisti A, Bowman V, Efimov A, Kurochkina L, Chipman P, Mesyanzhinov V, Rossmann M

EMDB-1333: 
The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities.
Method: single particle / : Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Muhlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ

EMDB-1334: 
The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities.
Method: single particle / : Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Muhlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ

EMDB-1335: 
The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities.
Method: single particle / : Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Muhlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ

EMDB-1336: 
The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities.
Method: single particle / : Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Muhlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ

EMDB-1337: 
The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities.
Method: single particle / : Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Muhlenhoff M, Gerardy-Schahn R, Scholl D, Molineux IJ

EMDB-1287: 
Asymmetric binding of transferrin receptor to parvovirus capsids.
Method: single particle / : Hafenstein S, Palermo LM, Xiao C, Kostyuchenko VA, Morais M, Nelson CDS, Chipman PR, Bowman VD, Battisti AJ, Parrish CR, Rossmann MG

EMDB-1288: 
Asymmetric binding of transferrin receptor to parvovirus capsids.
Method: single particle / : Hafenstein S, Palermo LM, Xiao C, Kostyuchenko VA, Morais M, Nelson CDS, Chipman PR, Bowman VD, Battisti AJ, Parrish CR, Rossmann MG

EMDB-1260: 
Structural changes of bacteriophage phi29 upon DNA packaging and release.
Method: single particle / : Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG

EMDB-1265: 
Structural changes of bacteriophage phi29 upon DNA packaging and release.
Method: single particle / : Xiang Y, Morais MC, Battisti AJ, Grimes S, Jardine PJ, Anderson DL, Rossmann MG

EMDB-1166: 
Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN.
Method: single particle / : Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG
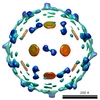
EMDB-1167: 
Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN.
Method: single particle / : Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao C, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
